Everything Eats Everyone
Left: Figure 1. Changes in soil structure are clearly visible afterFolsomia spp. (Collembola) where kept within a mesocosm of defaunated sieved soil for three months.
One of the most important components of the soil is the biology within it; this abundant and diverse group is the driver of nutrient cycling, decomposition, bioturbation and pedogenesis (Figure 1) within soil ecosystems. However due to the opacity of the soil, small size of the organisms involved and the diversity of species, understanding the interactions that are occurring is very difficult. It is because of these difficulties that the soil food web has often been referred to as a “black box” (Bonkowski et al., 2009), a “poor man’s tropical rainforest” (Giller, 1996) and an “enigma” (Anderson, 1975).
The large diversity of soil fauna has proved to be a headache for taxonomists, with no one able to be an expert in all soil fauna (Figure 2). This has led to a great deal of specialisation, with labs focusing on one particular taxon e.g. nematodes, or oribatid mites, or Collembola; therefore only a few investigate the soil food web as a whole.
Right: Figure 2. Small sample of fauna diversity from a Tullgren funnel extraction of one kilogram of soil.
Concentrating on the pathway of consumption through the soil food web is one method that can be utilised to investigate the whole ecosystem. There are a number of methods that can be used to understand feeding preferences, these are either observational (i.e. direct observation, culturing or gut content analysis), inference (mouthpart morphology, presence of certain digestive enzymes), or biochemical reactions (PLFAs or stable isotope analysis).
Stable isotopes can be utilised in two ways, firstly to ascertain the individual feeding interactions occurring. This can be accomplished by tracing an enriched isotope through different organisms to prove consumption (e.g. Crotty et al., 2011 and Crotty et al., 2012). This work showed that the Entomobryomorpha (Collembola) consumed traceable quantities of stable isotopically enriched bacteria and protozoa. This enrichment was also visible in the next trophic level, with Mesostigmatid mites also obtaining above background levels of enrichment. However, how these predators obtain enrichment, is ambiguous as they could be consuming the isotopically enriched source (bacteria or protozoa), or they could be consuming members of the mesofauna that previously consumed the enriched food source.
Secondly, stable isotopes can be used to try to understand the “bigger picture” of what is occurring within the soil food web. This can be achieved by comparing the differences between habitats and the naturally occurring stable isotope signatures of the organisms inhabiting them (Figure 3). The stable isotope signature of an organism is representative of the habitat it’s occupying (δ13C) whilst also representing the trophic level it’s feeding at (δ15N).
Left: Figure 3. Isotopic composition (δ13C and δ15N) of soil fauna within a grassland (black circles; blue labels) and a woodland (open circles; red labels) habitat of the same soil type, with the soil stable isotope signature for each habitat set to zero and all the other results calibrated accordingly (for abbreviations see Table 2 of Crotty et al., 2014).
In a recent paper by Crotty et al., (2014), isotope results indicate a divergence of feeding channels dependent on ecosystem type, even though previously the two ecosystems had been the same, prior to a change in management which converted a grassland to a woodland, over twenty-five years ago. In Figure 3, there is a visible separation between the grassland and woodland invertebrates for their δ15N values, suggesting that similar organisms are acting in different ways depending on the ecosystem they’re populating.
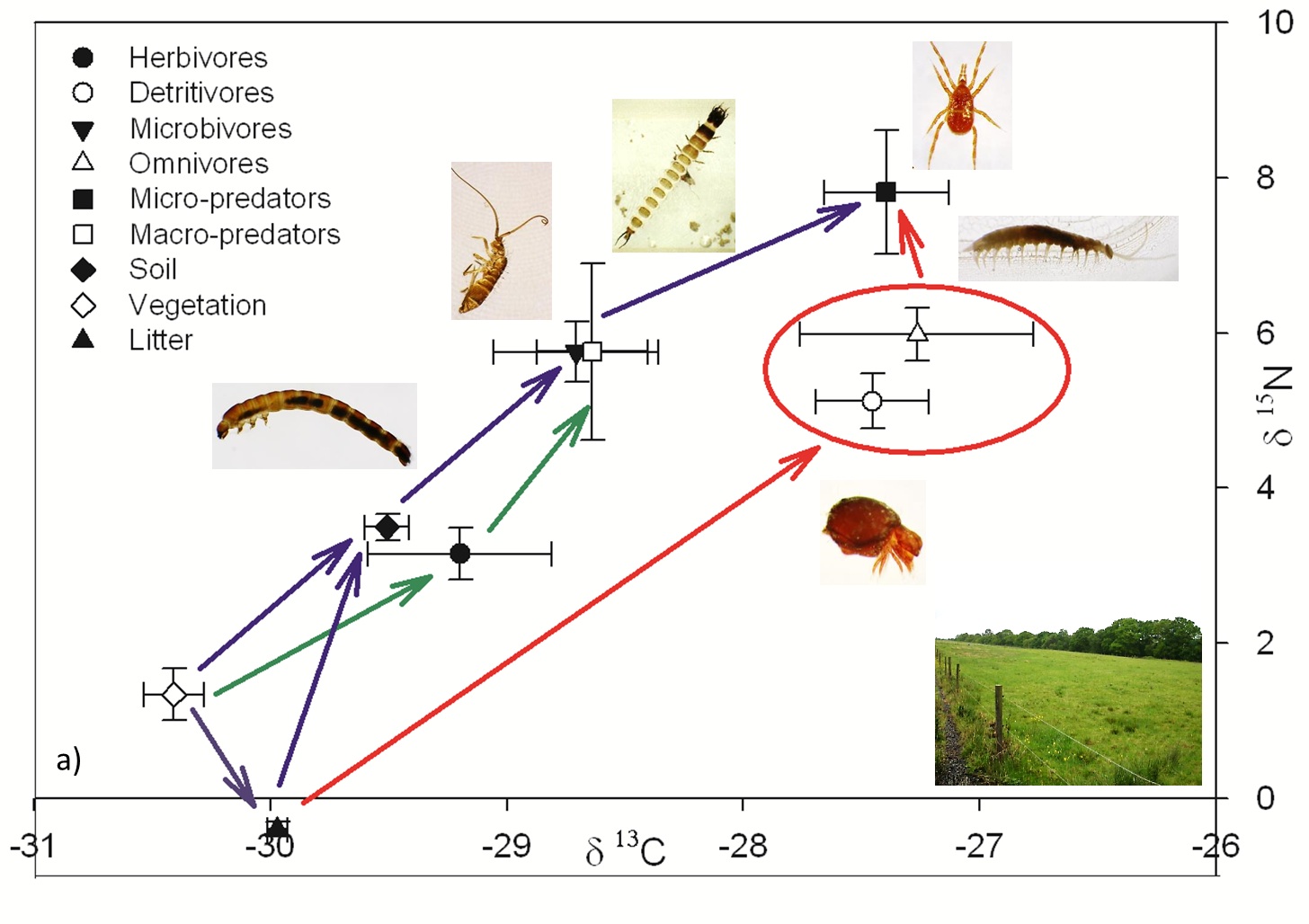
Further investigation concentrated on the differences in energy pathway within the soil food web between these two ecosystems. The grassland habitat promoted three defined feeding pathways (Figure 4a), whereas these pathways where more ambiguous in the woodland (Figure 4b), see Crotty et al., (2014) for a more detailed review. The isotope signatures of the taxa acting as “top predator” in the two ecosystems were very different. The micropredators appeared to exist at a higher trophic level than the macropredators in the grassland, likely to be because the prey organisms of the macropredators, where operating at a lower trophic level, than the micropredators prey.
Figure 4a.
Figure 4b.
Above: Figure 4a and 4b. Isotopic composition (δ13C and δ15N) of grouped “feeding guilds” for the (A) grassland habitat and (B) woodland habitat (Crotty et al., 2014). Arrows represent different feeding pathways – blue microbial, green herbivory, and red detritivore.
Our difficulty understanding the soil food web increases, largely because there are no direct predator-prey relationships occurring, with the majority of organisms eating a range of other organisms. This is probably due to the environment they reside in, where prey species that are in close proximity may be invisible as they are located in an adjacent soil pore that is not easily accessible. Unlike in aquatic environments (where food web studies originated), the soil food web is one where the mixing of food and waste is commonplace. All “waste” within the soil is utilized by other organisms; microbes directly break it down, or mesofauna consuming the microbiota and waste, recycling the nutrients. Intraguild predation and carrion consumption increase the potential mixing of isotopic signatures. Together, this leads to a dilution of the distinctiveness of trophic levels within the soil system. Further work is still needed to assess the contribution of these different feeding channels within nutrient cycling, to gain a better understanding of how to promote a healthy soil.
References:
Anderson J.M. (1975). The Enigma of Soil Animal Species. In: Progress in Soil Zoology: Proceedings of the Fifth International Colloquium on Progress in Soil Zoology (ed. Vanek J). Prague Academia: The Hague, pp. 51-58.
Bonkowski M., Villenave C. & Griffiths B. (2009). Rhizosphere fauna: the functional and structural diversity of intimate interactions of soil fauna with plant roots. Plant Soil, 321, 213-233.
Crotty F.V., Blackshaw R.P., Adl S.M., Inger R. & Murray P.J. (2014). Divergence of feeding channels within the soil food web determined by ecosystem type. Ecology and Evolution, 4, 1-13.
Crotty F.V., Adl S.M., Blackshaw R.P. & Murray P.J. (2012). Protozoan pulses unveil their pivotal position within the soil food web. Microb. Ecol., 63, 905-918.
Crotty F.V., Blackshaw R.P. & Murray P.J. (2011). Tracking the flow of bacterially derived 13C and 15N through soil faunal feeding channels. Rapid Commun. Mass Spectrom., 25, 1503-1513.
Giller P.S. (1996). The diversity of soil communities, the 'poor man's tropical rainforest'. Biodivers. Conserv., 5, 135-168.